“In Silico: in or on a computer : done or produced by using computer software or simulation“
https://www.merriam-webster.com/dictionary/in%20silico
I won’t go into too much detail regarding the Drosten PCR test as it was covered beautifully by people much better suited than I (linked below), but I would be remiss if I did not mention it at all. Drosten was instrumental in the fraud that was “SARS-COV-1” and naturally, his experience in regards to manipulating the masses with pseudoscience was put to good use with “SARS-COV-2.” He has so much experience with these “viruses,” he was able to create a “diagnostic” PCR test in silico in the absence of any “SARS-COV-2 isolates” based primarily off of social media reports. That takes talent folks. Highlights below:
Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR
Background
“The ongoing outbreak of the recently emerged novel coronavirus (2019-nCoV) poses a challenge for public health laboratories as virus isolates are unavailable while there is growing evidence that the outbreak is more widespread than initially thought, and international spread through travellers does already occur.
Aim
We aimed to develop and deploy robust diagnostic methodology for use in public health laboratory settings without having virus material available.
Methods
Here we present a validated diagnostic workflow for 2019-nCoV, its design relying on close genetic relatedness of 2019-nCoV with SARS coronavirus, making use of synthetic nucleic acid technology.
Results
The workflow reliably detects 2019-nCoV, and further discriminates 2019-nCoV from SARS-CoV. Through coordination between academic and public laboratories, we confirmed assay exclusivity based on 297 original clinical specimens containing a full spectrum of human respiratory viruses. Control material is made available through European Virus Archive – Global (EVAg), a European Union infrastructure project.”
“A novel coronavirus currently termed 2019-nCoV was officially announced as the causative agent by Chinese authorities on 7 January. A viral genome sequence was released for immediate public health support via the community online resource virological.org on 10 January (Wuhan-Hu-1, GenBank accession number MN908947 [2]), followed by four other genomes deposited on 12 January in the viral sequence database curated by the Global Initiative on Sharing All Influenza Data (GISAID). The genome sequences suggest presence of a virus closely related to the members of a viral species termed severe acute respiratory syndrome (SARS)-related CoV, a species defined by the agent of the 2002/03 outbreak of SARS in humans [3,4]. The species also comprises a large number of viruses mostly detected in rhinolophid bats in Asia and Europe.”
“Among the foremost priorities to facilitate public health interventions is reliable laboratory diagnosis. In acute respiratory infection, RT-PCR is routinely used to detect causative viruses from respiratory secretions. We have previously demonstrated the feasibility of introducing robust detection technology based on real-time RT-PCR in public health laboratories during international health emergencies by coordination between public and academic laboratories [6–12]. In all of these situations, virus isolates were available as the primary substrate for establishing and controlling assays and assay performance.
In the present case of 2019-nCoV, virus isolates or samples from infected patients have so far not become available to the international public health community. We report here on the establishment and validation of a diagnostic workflow for 2019-nCoV screening and specific confirmation, designed in absence of available virus isolates or original patient specimens. Design and validation were enabled by the close genetic relatedness to the 2003 SARS-CoV, and aided by the use of synthetic nucleic acid technology.
Methods
Clinical samples and coronavirus cell culture supernatants for initial assay evaluation
Cell culture supernatants containing typed coronaviruses and other respiratory viruses were provided by Charité and University of Hong Kong research laboratories. Respiratory samples were obtained during 2019 from patients hospitalised at Charité medical centre and tested by the NxTAG respiratory pathogen panel (Luminex, S´Hertogenbosch, The Netherlands) or in cases of MERS-CoV by the MERS-CoV upE assay as published before [10]. Additional samples were selected from biobanks at the Rijksinstituut voor Volksgezondheid en Milieu (RIVM), Bilthoven, at Erasmus University Medical Center, Rotterdam, at Public Health England (PHE), London, and at the University of Hong Kong. Samples from all collections comprised sputum as well as nose and throat swabs with or without viral transport medium.
Faecal samples containing bat-derived SARS-related CoV samples (identified by GenBank accession numbers) were tested: KC633203, Betacoronavirus BtCoV/Rhi_eur/BB98–98/BGR/2008; KC633204, Betacoronavirus BtCoV/Rhi_eur/BB98–92/BGR/2008; KC633201, Betacoronavirus BtCoV/Rhi_bla/BB98–22/BGR/2008; GU190221 Betacoronavirus Bat coronavirus BR98–19/BGR/2008; GU190222 Betacoronavirus Bat coronavirus BM98–01/BGR/2008; GU190223, Betacoronavirus Bat coronavirus BM98–13/BGR/2008.
All synthetic RNA used in this study was photometrically quantified.”
Real-time reverse-transcription PCR
“A 25 μL reaction contained 5 μL of RNA, 12.5 μL of 2 × reaction buffer provided with the Superscript III one step RT-PCR system with Platinum Taq Polymerase (Invitrogen, Darmstadt, Germany; containing 0.4 mM of each deoxyribont triphosphates (dNTP) and 3.2 mM magnesium sulphate), 1 μL of reverse transcriptase/Taq mixture from the kit, 0.4 μL of a 50 mM magnesium sulphate solution (Invitrogen), and 1 μg of nonacetylated bovine serum albumin (Roche). Primer and probe sequences, as well as optimised concentrations are shown in Table 1. All oligonucleotides were synthesised and provided by Tib-Molbiol (Berlin, Germany). Thermal cycling was performed at 55 °C for 10 min for reverse transcription, followed by 95 °C for 3 min and then 45 cycles of 95 °C for 15 s, 58 °C for 30 s.”
“The intended cross-reactivity of all assays with viral RNA of SARS-CoV allows us to use the assays without having to rely on external sources of specific 2019-nCoV RNA.”
Results
“Before public release of virus sequences from cases of 2019-nCoV, we relied on social media reports announcing detection of a SARS-like virus. We thus assumed that a SARS-related CoV is involved in the outbreak. We downloaded all complete and partial (if > 400 nt) SARS-related virus sequences available in GenBank by 1 January 2020. The list (n = 729 entries) was manually checked and artificial sequences (laboratory-derived, synthetic, etc), as well as sequence duplicates were removed, resulting in a final list of 375 sequences. These sequences were aligned and the alignment was used for assay design (Supplementary Figure S1). Upon release of the first 2019-nCoV sequence at virological.org, three assays were selected based on how well they matched to the 2019-nCoV genome (Figure 1). The alignment was complemented by additional sequences released independently on GISAID (https://www.gisaid.org), confirming the good matching of selected primers to all sequences. Alignments of primer binding domains with 2019-nCoV, SARS-CoV as well as selected bat-associated SARS-related CoV are shown in Figure 2.
Assay sensitivity based on SARS coronavirus virions
To obtain a preliminary assessment of analytical sensitivity, we used purified cell culture supernatant containing SARS-CoV strain Frankfurt-1 virions grown on Vero cells. The supernatant was ultrafiltered and thereby concentrated from a ca 20-fold volume of cell culture supernatant. The concentration step simultaneously reduces the relative concentration of background nucleic acids such as not virion-packaged viral RNA. The virion preparation was quantified by real-time RT-PCR using a specific in vitro-transcribed RNA quantification standard as described in Drosten et al. [8].”
Discrimination of 2019 novel coronavirus from SARS coronavirus by RdRp assay
“Following the rationale that SARS-CoV RNA can be used as a positive control for the entire laboratory procedure, thus obviating the need to handle 2019-nCoV RNA, we formulated the RdRp assay so that it contains two probes: a broad-range probe reacting with SARS-CoV and 2019-nCoV and an additional probe that reacts only with 2019-nCoV. By limiting dilution experiments, we confirmed that both probes, whether used individually or in combination, provided the same LOD for each target virus. The specific probe RdRP_SARSr-P2 detected only the 2019-nCoV RNA transcript but not the SARS-CoV RNA.”
“To show that the assays can detect other bat-associated SARS-related viruses, we used the E gene assay to test six bat-derived faecal samples available from Drexler et al. [13] und Muth et al. [14]. These virus-positive samples stemmed from European rhinolophid bats. Detection of these phylogenetic outliers within the SARS-related CoV clade suggests that all Asian viruses are likely to be detected. This would, theoretically, ensure broad sensitivity even in case of multiple independent acquisitions of variant viruses from an animal reservoir.”
Cross-reactivity with other coronaviruses
“Cell culture supernatants containing all endemic human coronaviruses (HCoV)‑229E, ‑NL63, ‑OC43 and ‑HKU1 as well as MERS-CoV were tested in duplicate in all three assays (Table 2). For the non-cultivable HCoV-HKU1, supernatant from human airway culture was used. Viral RNA concentration in all samples was determined by specific real-time RT-PCRs and in vitro-transcribed RNA standards designed for absolute quantification of viral load. Additional undiluted (but not quantified) cell culture supernatants were tested as summarised in Table 2. These were additionally mixed into negative human sputum samples. None of the tested viruses or virus preparations showed reactivity with any assay.”
Discussion
“The present report describes the establishment of a diagnostic workflow for detection of an emerging virus in the absence of physical sources of viral genomic nucleic acid. Effective assay design was enabled by the willingness of scientists from China to share genome information before formal publication, as well as the availability of broad sequence knowledge from ca 15 years of investigation of SARS-related viruses in animal reservoirs.”
“Technical qualification data based on cell culture materials and synthetic constructs, as well as results from exclusivity testing on 75 clinical samples, were included in the first version of the diagnostic protocol provided to the WHO on 13 January 2020. “
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6988269/#!po=0.746269
In Summary:
- No “viral isolates” were available at the time Drosten created his PCR test
- The aim of his study was to develop a test without needing “virus” material
- The workflow was based on the “close genetic-relatedness” between “SARS-COV-1” and “SARS-COV-2” (only 79% which is really not close at all) with the use of SYNTHETIC nucleic acid technology
- They based assay exclusivity off of 297 clinical samples containing a range of respiratory “viruses” but none with “SARS-COV-2”
- The genome sequence supplied by the Chinese SUGGESTED a “virus” related to “SARS-COV-1”
- In their previous test developments, “viral isolates” were available as the primary substrate for establishing and controlling assays and assay performance
- Since no “virus isolates” were available this time around, they decided that the presumed relation to “SARS-COV-1” was enough to develop a reliable test with the use of synthetic nucleic acid technology
- Cell culture supernatant for different “viruses” were provided by the Charite and various other sources
- Samples were from sputum as well as nose and throat swabs with or without viral transport media
- They also used fecal samples supposedly containing Bat-related “Coronaviruses”
- Synthetic RNA was used in the study
- During RT-PCR, various chemicals were used along with nonacetylated bovine serum albumin
- All oligonucleotides were synthesized
- The RT-PCR tests were run to 45 Cycles (remember, according to Fauci, anything above 35 is just dead nucleotides)
- The intended cross-reactivity of all assays to “SARS-COV-1” somehow meant they did not need any “SARS-COV-2” isolates
- They relied on social media reports and assumed from those that a “Coronavirus” related to “SARS-COV-1” was the culprit
- Numerous different “SARS-COV-1” genomes (375) were aligned and used to design the assays for “SARS-COV-2”
- 3 assays were chosen based on how well they matched up with the “SARS-COV-2” genome
- They used “purified SARS-COV-1” grown on Vero Cells to test analytical sensitivity
- They ultrafiltered the cell culture supernatant to REDUCE (not eliminate) the background nucleic acids such as that of not virion-packaged “viral” RNA
- They “rationalized” (cough assumed cough) that “SARS-COV-1” could act as a positive control for “SARS-COV-2” in all tests
- They determined that the E Gene assay can detect all Asian “viruses”
- They also admit HCoV-HKU1 is non-cultivable
- For some reason, they mixed “viral” culture supernatant with “non-viral” human sputum samples to test vs just using the unaltered human samples
- The originally submitted technological qualifications were based on cell culture materials and synthetic constructs
- The study itself was peer-reviewed and accepted in less than 24 hours
For an in-depth breakdown of this the Drosten PCR fraud, read the Corman-Drosten Review Report:
Review report Corman-Drosten et al. Eurosurveillance 2020
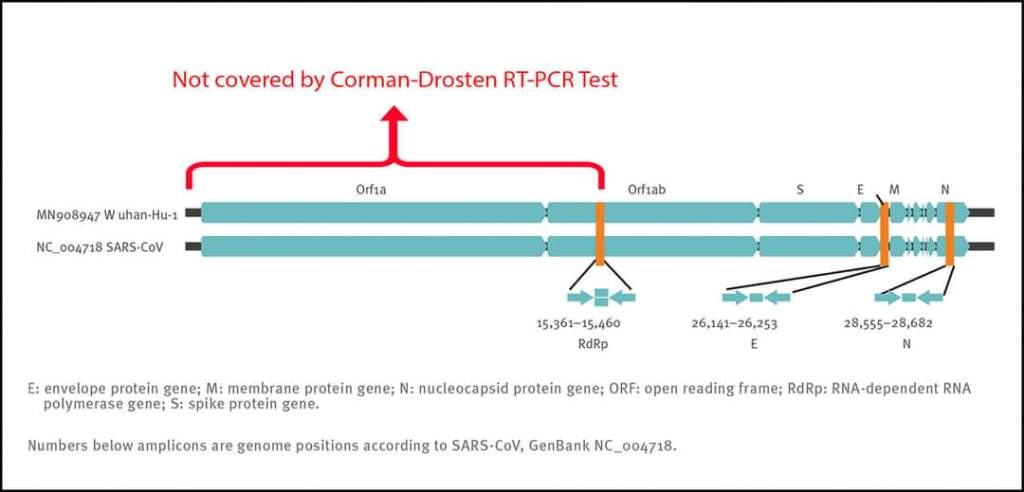
This is a brief list of what was uncovered:
“In November 2020, 22 internationally renowned scientists conducted an external peer review of Corman’s paper to independently assess its quality and accuracy. In this publication, Borger, et al. [6] concluded that the article published without guarantees in Eurosurveillance contains nine serious scientific errors and three minor inaccuracies.
The detailed explanation of these scientific errors exceeds the
remit of this editorial, which is why they are only listed below.
- Extremely high concentrations of primers, DNA polymerase and magnesium sulfate. This leads to an increase in nonspecific binding, amplification, and lack of specificity to identify the SARS-CoV-2 virus.
- Non-specific primers (oscillating letters) that could give rise to various sequences of forward primers and as many inverse ones that are not related at all to SARS-CoV-2, so the test is not a specific tool for its diagnosis.
- The test cannot discriminate between the complete virus (infectious) and the viral fragments.
- A difference of 10 °C with respect to the annealing temperature of the first pair of RdRp primers (direct and reverse) when it should be 2 °C.
- The genes chosen were wrong because:
- They did not represent the entire length of the virus.
- The E gene is nonspecific and is present in all coronaviruses.
- The N gene that at least ensured that it was a SARS-1 or SARs-CoV2 was removed by the WHO from the protocol due to lack of sensitivity.
- The RdPd gene proposed by Corman, et al. [4] contains too many oscillating letters so that 2 forward primers, 4 different reverse primers and 8 different probes could be synthesized, which provides excessive variability from the point of view of commercial tests to ensure its specificity.
- The PCR products have not been validated at the molecular level.
- The PCR test does not contain a single positive control to evaluate its specificity for SARS-CoV-2 or a negative control to exclude the presence of other coronaviruses, which makes the test unsuitable as a specific diagnostic tool to identify SARS-CoV-2. Virus
- The number of cycles is not specified for a test to be positive. Later, the WHO recommended between 40 and 45 cycles, which is totally wrong from a scientific point of view.”
And just in case you thought the CDC was above creating a PCR test without any “virus” available, know you would be wrong.
This is from the FDA emergency use authorization of the CDC’s PCR test used in the USA:
“Since no quantified virus isolates of the 2019-nCoV are currently available, assays designed for detection of the 2019-nCoV RNA were tested with characterized stocks of in vitro transcribed full length RNA (N gene; GenBank accession: MN908947.2) of known titer (RNA copies/µL) spiked into a diluent consisting of a suspension of human A549 cells and viral transport medium (VTM) to mimic clinical specimen.”
https://www.fda.gov/media/134922/download
Is it any wonder why both the Drosten and CDC PCR tests performed horribly regarding false-positives?
“The E Charité and N2 US CDC assays were positive for all specimens, including negative samples and negative controls (water). These false-positive results were explored (details below), but the sensitivity of these assays was not further assessed.”
“It is worth noting that the Charité assay was the first to be published at the early stage of the pandemic [9] and has been widely used worldwide [8].”
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7355678/

So it appears there is no need to have “viruses” available to create and validate tests anymore. Drosten and others can get on Twitter, read some social media reports, and then come up with a test for a “novel virus” on their computers without ever needing the actual “virus” present. They can just use the old not-as-closely-related-as-they-would-like-you-to-believe “viruses” as a stand-in. Pretty neat trick with how everything, from the “novel virus” itself to the the test to detect it, are all computer-based. Those virologists sure dodged a bullet with their molecular tricks and consensus computer algorithms as there is no need for the gold standard of a purified/isolated “virus.” Social media driven computer-based synthetic creations are all the rage these days.

In Italia continuano tenere banco la falsa informazione del Parlamento circa il virus con la convinzione che puoi infettarti da persona a persona. Hanno forse effettuato accordi lucubri o sotto pressione con le case farmaceutiche, quindi minacciando il popolo, se non ti vaccini ti isolo dalla società anzi non ti faccio lavorare e tutto questo sta avvenendo dopo 3 inverni. Il tutto con la compiacenza della Magistratura? Altro che Norimberga 2…
LikeLiked by 1 person
Your response in English:
“In Italy, the false information of the Parliament about the virus continues with the belief that you can get infected from person to person. They may have made lucrative or pressured agreements with pharmaceutical companies, so by threatening the people, if you don’t get vaccinated I isolate you from society or rather I don’t make you work and all this is happening after 3 winters. All with the complacency of the judiciary? Other than Nuremberg 2 …”
My response in English:
I agree completely. Deals have been made with the pharmaceutical companies throughout the world in an attempt to force and/or coerce people into unnecessary and dangerous medical interventions as well as to take power away from the people. We need everyone to wake up and stand together in order to demand this madness stops now.
My response in Italian:
“Sono completamente d’accordo. Sono stati presi accordi con le aziende farmaceutiche di tutto il mondo nel tentativo di costringere e/o costringere le persone a interventi medici inutili e pericolosi, nonché di togliere potere alle persone. Abbiamo bisogno che tutti si sveglino e stiano uniti per chiedere che questa follia finisca ora.”
LikeLike